Chapter 5 Coding Handbook
5.1 Justification
The practice of science requires special care to ensure integrity. Not only do we want to know our results are correct, we need to show outside collaborators, institutions, publishers, and funders. The standards for these groups are also rising, particularly in the areas of data and code. Excerpts from the Fostering Integrity in Research (2018) checklists for researchers, journals, and research sponsors:
Researchers:
- Develop data management and sharing plan at the outset of a project.
- Incorporate appropriate data management expertise in the project team.
- Understand and follow data collection, management, and sharing standards, policies, and regulations of the discipline, institution, funder, journal, and relevant government agencies.
Journals:
- Provide a link to data and code that support articles, and facilitate long-term access.
- Require full descriptions of methods in method sections or electronic supplements.
Research sponsors:
- Develop data and code access policies for extramural grants appropriate to the research being funded, and make fulfillment of these policies a condi- tion of future funding.
- Cover the costs borne by researchers and institutions to make data and code available.
- Practice transparency of data and code for intramural programs.
- Promote responsible sharing of data in areas such as clinical trials.
One of the main reasons research has changed with respect to ensuring integrity in the last few decades is the increasing role of data and computer software. New norms, standards, and training are required, and new opportunities for communication and reuse are available.
5.2 Coding Culture
As with any high-stakes endeavor, it’s important to think about how our treatment of each other contributes to success. Software development depends on accuracy, interdependence, and requires human judgment. Culture can therefore greatly impact productivity and resiliency.
5.2.1 Cultural practices
Be open with your code, and your understanding. Coding productivity depends on information. Passing the information as quickly and openly as possible will help overcome this and facilitate progress. The communication phenomenon has been known since the 70s yet it is easy to forget.
Be charitable with your feedback. No error is so obvious that even the most experienced programmer won’t make it from time to time. Break your PR reviews into demonstrable chunks that you can prove with a code snippet. Don’t make sweeping or vague judgements.
Be thick-skinned. Code has a way of seeming absolute and damning when you get it wrong. On the other hand, that is its nature. Any error will feel that way, and everyone makes mistakes.
5.3 Complexity
The art of programming is the art of organizing complexity. — Edsger W. Dijkstra
Programs tend to be difficult to understand. They are written by someone with roughly the same capacity for complexity as the reader, but with the advantage of having written it. This person will usually write to the limit of their own understanding because we write competitive programs that are as sophisticated and full-featured as possible.
There are strong incentives in all of computing to write complex code. There are also a limitless number of ways to write code that are functionally identical to each other.
It’s also important to remember that complexity is a force of nature. Once enough possible states of a program have been achieved that are difficult to characterize – which is easy to do – it becomes impossible for any human brain to understand completely. This doesn’t happen for all programs, but due to combinatorics, the point at which an application becomes complex can easily sneak up on the author.
5.4 Why writing code is easier than reading it
The process of understanding a code practically involves redoing it. — John von Neumann
Take a function in a large codebase. The person who wrote it understands:
- The expected – or possible – range of inputs for this particular application. (Number of possible arguments – values – can easily be in the trillions for a simple function.) Possible range will often depend on the entire rest of the codebase and will usually be implicit.
- How often the function is called at runtime (Note: This is different from the number of times it is referenced in the codebase.)
- The intentions of the code. This can be different from what it does and is simpler to understand. (Usually intention vs reality is cleared up with comments.)
- The narrative of the code. The history of a codebase is a powerful mnemonic device. “We wrote this because there was an issue in January. There are three other places this functionality is handled.”
All of this asymmetry between the author and the reader is in addition to the raw size of the source code. In other words, these depend on the combinatorial explosion of interconnected components.
5.4.1 Science and Software
Some coding principles are less important in science because:
- Scientific applications are more mathematical. You can reason about the range of values more easily.
- They can be very short.
- They are often meant to be run one way. For example, a Jupyter notebook that is intended to run in the order the cells are in on the page.
Some coding principles are more important in science because:
- Scientific applications are meant to be read. They are intended to teach and be verified.
- They are meant to be open-source, and reused.
- They are at the forefront of human understanding. Extra complexity is detrimental.
- They are sensitive to error. The stakes are high.
5.5 Indirection, Abstraction, and Generalization
Indirection, abstraction, and generalization are three closely-related concepts in programming.
Indirection is the most general in that it refers to any symbolic representation of a process in the place of the process itself. A function calling another function, for instance.
Indirection can reduce complexity, and multiply the number of cases a piece of code can handle, and be a source of complexity itself.
The so-called Fundamental Theorem of Software Engineering, attributed to David J. Wheeler, is: “We can solve any problem by introducing an extra level of indirection. (Except for the problem of too many levels of indirection.)”
Abstraction is also very general but refers to the process of removing details that are not relevant to some concept one is trying to model.
Finally, generalization is very similar to abstraction with the connotation of combining the functionality of several similar pieces of code into one, usually parameterized, copy.
Thinking about how accurately, simply, and powerfully your code represents what is being modeled is important because it makes your code more useful, understandable, and because it becomes more mathematical: You’ve distilled a model to its essence.
5.5.1 Example: The Weasel Algorithm
from random import choice, random
charset = 'abcdefghijklmnopqrstuvwxyzABCDEFGHIJKLMNOPQRSTUVWXYZ '
target = list("METHINKS IT IS LIKE A WEASEL")
# create a random parent
parent = [choice(charset) for _ in range(28)]
while parent != target:
# calculate how fast to mutate depending on how close we are to the target.
rate = 1-((28 - (sum(t == h for t, h in zip(parent, target))
)) / 28 * 0.9)
# initialize ten copies of the parent, randomly mutated.
mut1 = [(ch if random() <= rate else choice(charset)) for ch in parent]
mut2 = [(ch if random() <= rate else choice(charset)) for ch in parent]
mut3 = [(ch if random() <= rate else choice(charset)) for ch in parent]
mut4 = [(ch if random() <= rate else choice(charset)) for ch in parent]
mut5 = [(ch if random() <= rate else choice(charset)) for ch in parent]
mut6 = [(ch if random() <= rate else choice(charset)) for ch in parent]
mut7 = [(ch if random() <= rate else choice(charset)) for ch in parent]
mut8 = [(ch if random() <= rate else choice(charset)) for ch in parent]
mut9 = [(ch if random() <= rate else choice(charset)) for ch in parent]
mut10 = [(ch if random() <= rate else choice(charset)) for ch in parent]
# put mutants in an array
copies = [mut1, mut2, mut3, mut4, mut5, mut6, mut7, mut8, mut9, parent]
# pick most fit parent from the beginning of the list
parent1 = max(copies[:4], key=lambda trial: sum(t == h for t, h in zip(trial, target)))
# pick most fit parent from the end of the list
parent2 = max(copies[4:], key=lambda trial: sum(t == h for t, h in zip(trial, target)))
# choose a place in the "genome" to split between the two parents.
place = choice(range(28))
mated = [parent1, parent2, parent1[:place] + parent2[place:], parent2[:place] + parent1[place:]]
# choose most fit amongst the parents and two progeny
parent = max(mated, key=lambda trial: sum(t == h for t, h in zip(trial, target)))
print(''.join(parent))
print('Success! \n', ''.join(parent))This code has several problems.
- There’s a lot of duplication. Initializing the array requires as many lines as there are elements in the array.
sum(t == h for t, h in zip(trial, target))is copied anywhere it is needed, etc. - It only works for specific lengths of
targetandcopiesbecause a user would need to modify code instead of changing a parameter. Literal values like 4 and 28 are used instead of variables (hardcoding). - It’s not conceptual. If it weren’t for the comments, it would be difficult to tell what the author is getting at. What does 28 mean in this context? Is it the same as other 28s?
- It would be difficult to maintain (particularly if this style was used in a large program). Will the program still work if we change some of it? Is there an error in some of the duplicated code? How do we add functionality without adding more duplication?
All of these are problems that can be solved by generalization.
This code was adapted from a much more general version at Rosetta Code. In some ways the less general version is easier to understand. Your eye doesn’t need to jump around as much to see what is going on. Usually, though, the more general code is preferred.
How a particular program should be written is a judgment call based on its size, predicted longevity, and what is most clear to readers. If you’re starting to lose productivity or bugs are difficult to fix, it might be time to generalize. As with almost any engineering topic, generalization is a tradeoff and can be taken too far.
5.6 Debugging
Debugging and experimentation are fundamentally the same. Debugging is done by isolating variables to identify the cause of a problem. It is comparing the results of two runs of a program with one variable changed. If one run reproduces a bug and the other doesn’t, you can usually conclude the value of the variable is the cause. (“Variable” may not be a literal variable. It may be a short section of code, or input.)
Modular, functional code is easier to debug than the opposite: arbitrarily interconnected code. The reason for this is it’s easier to understand, and it’s easier to “change one thing” and understand the outcome. Code that is easy to debug is also generally modular, functional, and testable.
5.7 Functional Programming
The term “functional” comes from Functional Programming, which is a discipline in which:
- Functions always return the same output for an input. For instance:
Always returns the corresponding x + 2 for every x no matter the context and across time.
Does not.
- Functions have no side-effects. The function can’t modify any variables outside itself.
Modifies y, which is outside the scope of the function f. State applies to external systems as well. Modifying a database, for instance, counts as out-of-scope, and can affect future runs of a function with the same input.
These properties guarantee that a program is referentially transparent. You can easily modify it because you can replace any instance of a function with a value, and you can move functions without concern that their behavior will change. Additionally, variables that can be changed by many different functions, in the worst case global mutable variables, add complexity to programs. This is analogous to running an experiment where variables can’t be controlled because the state of the program generally involves variables that could be at any state at any time and may radically change the behavior of the program. A function in functional programming (also known as a pure function) can be tested completely by changing its arguments.
The benefits of functional programming are closely related to modularity. Functional programs are modular in that every function encapsulates some functionality and has a well-defined interface, the function signature.
5.8 Unit Testing
Unit testing is a technique for verifying a codebase by writing sample input and expected output for a number of its functions. These tests – usually structured as functions themselves – are binary. They either run to completion, or they throw an exception. Failure to run represents failure of the test.
Unit tests are generally run without input. The input to the function being tested is written as literals (3, "blue", [6, 7, 8]) within the test function, or globally for several tests to use. Input to a test, or variables and environment necessary for the function to run is called a fixture.
A test suite is the set of all tests of a codebase. Usually test suites are run in their entirety, giving a simple output with the percentage of tests that passed. The codebase (usually called a “build” in this context) is said to be passing or failing.
Unit testing serves two major purposes.
- A Unit test is a declaration by the author that an input/output pair is “correct.” They represent what the function should do. A programmer could write a unit test
test_addof the functionadd:
def add(a, b):
"""One of the four basic arithmetic operations. Takes two numbers -- a and b -- and returns a + b."""
return a + b + 1
def test_add():
assert add(1, 1) == 3The test verifies that the input/output pair (a = 1, b = 1)/3 is consistent with the function add, but does not validate that it is functioning properly (at least according to the way add has been described).
A valid test:
would uncover the fact that add was either written incorrectly, or was recently broken.
When writing tests, it’s good to check yourself by avoiding the output of the function as it is written. Take input/output pairs from another, reliable, source or think about the problem and write what you believe is the correct output. A programmer who runs add and then writes a test with (a = 1, b = 1)/3 would be perpetuating the error instead of correcting it.
- Unit tests verify that changes to a codebase don’t have unexpected effects. In other words, they help compare two versions of the code to show the functionality (set of input/output pairs) is the same.
This is helpful for refactoring or rewriting where many changes are being made and the application needs to be verified repeatedly.
5.8.1 Limitations to Unit Tests
Unit tests are not proofs. They test one input/output pair that stands for many pairs in the input/output space. It is always possible that one pair is not tested that will be critical to the functioning of the program, and the program does not handle it as intended.
It is important, therefore, that unit tests are written for representative pairs. For instance:
The test test_add_ones is representative of the space of three integers, but it ignores the conditional that modifies the behavior in the case of a = 3. Real-world examples will be much less obvious so this is common. The path may be one of thousands, buried in many tens of thousands of lines of code.
Because all of the paths that characterize the behavior of the function are not tested, this codebase could be said to have insufficient “coverage.” Unit testing libraries will usually be able to measure coverage, which is useful for finding these cases.
5.8.2 Testability
A function is easier to test when there’s a simple way to characterize its behavior with some inputs and expected outputs. This generally means small, easy-to-write input/output pairs. For example:
def f(i, j, s):
if i > j:
return s + " is above the line"
else:
return s + " is below the line"
def test_low_f():
assert f(2, 3, "test") == "test is below the line"
def test_high_f():
assert f(3, -1, "test") == "test is above the line"def g(i, j, s):
if i > j and is_thursday and urllib.request.urlopen("http://line.status.org") and random.randrange(1,10) > 2:
return "It's thursday and the line status is good and you're lucky."Function f: 1. Is purely functional. It doesn’t modify or depend on values outside its scope, and always returns the same values for the corresponding input.
f is easy to test. test_low_f and test_high_f cover both branches (the if and else) and characterize the behavior of f well. Note: It’s a judgment call whether or not enough of the input space has been tested. It’s easy to see in this case that all integers will behave predictably. This also doesn’t cover exceptions, which should generally be tested.
Function g: 1. Works differently depending on the day, the status of an external web site, and a random number. The behavior of g depends on a lot of values, and values that are outside the scope of the function. 2. Does not handle all values of its parameters.
g is not purely functional and very difficult to test. The state that would be needed to get a predictable output is difficult to prepare. (Functions should also almost never silently fail.)
This most commonly occurs in practice with large global, mutable variables and large objects in general, and the results of connections to external services. Sometimes this can’t be avoided, but structuring your program strategically can minimize the effects of state.
5.9 Modules and Modularity
The “messiness” of code is hard to quantify. Messy, difficult-to-understand code is sometimes called “spaghetti code” because connections between components are made from anywhere, to anywhere without much planning or structure. However:
- A complex codebase should have many connections such as function calls, imports, variable mutation etc.
- Where and when to make connections can be a matter of what “lens” through which you’re viewing the code. There isn’t an absolute objective standard.
Modularity is one path to clean, simple, maintainable code that is considered distinct from “spaghetti” and can be put in objective terms.
The word “module” has a specific meaning in certain programming languages. As a general term, it means a section of code that acts as a unit, usually on a particular topic or domain, that has a well-defined interface.
An interface is a boundary between components over which information is exchanged. The simplest well-defined interface is a function signature.
The “module” f has an interface that is the parameters a, b, and c. Any code that calls f needs to provide these parameters. They are defined explicitly in the codebase and enforced by the compiler. Variables within f can’t be accessed from outside f. In other words, a connection, or metaphorical spaghetti strand can’t be made to anything inside f.
Once a codebase is organized into modules, it becomes much simpler and easier to maintain. Modularity also contributes to “separation of concerns” one of the most important, if not the most important software principle. Software organized into concerns with interfaces in between is easier to reason about because the modules can be reasoned about, and modified, independently.
Returning to the evolutionary algorithm example, a more modular version might look like this:
"""Module for simulating evolution of strings."""
from random import choice, random
from functools import partial
def fitness(trial, target):
"""Compare a string with a target. The more matching characters, the higher the fitness."""
return sum(t == h for t, h in zip(trial, target))
def mutaterate(parent, target):
"""Calculate a mutation rate that shrinks as the target approaches."""
perfectfitness = float(len(target))
return (perfectfitness - fitness(parent, target)) / perfectfitness
def mutate(parent, rate, alphabet):
"""Randomly change characters in parent to random characters from alphabet."""
return [(ch if random() <= 1 - rate else choice(alphabet)) for ch in parent]
def mate(a, b):
"""
Split two strings in the center and return 4 combinations of the two:
1. a
2. b
3. beginning of a, then the end of b
4. beginning of b, then the end of a
"""
place = int(len(a)/2)
return a, b, a[:place] + b[place:], b[:place] + a[place:]
def evolve(seed, target_string, alphabet, population_size=100):
"""Randomly mutate populations of seed until it "evolves" into target_string."""
assert all([l in alphabet for l in target_string]), \
"Error: Target must only contain characters from alphabet."
assert len(seed) == len(target_string), \
"Error: Target and Seed must be the same length."
# For performance
target = list(target_string)
parent = seed
generations = 0
while parent != target:
rate = mutaterate(parent, target)
mutations = [mutate(parent, rate, alphabet) for _ in range(population_size)] + [parent]
center = int(population_size/2)
parent1 = max(mutations[:center], key=partial(fitness, target))
parent2 = max(mutations[center:], key=partial(fitness, target))
parent = max(mate(parent1, parent2), key=partial(fitness, target))
generations += 1
return generations
# Tests
import unittest
class TestEvolveMethods(unittest.TestCase):
def test_fitness(self):
self.assertEqual(fitness("abcd", "axxd"), 2)
self.assertEqual(fitness("abcd", "abcd"), 4)
self.assertEqual(fitness("abcd", "wxyz"), 0)
def test_mutaterate(self):
self.assertEqual(mutaterate("abcd", "wxyz"), 1)
self.assertEqual(mutaterate("abcd", "abce"), 0.25)
self.assertEqual(mutaterate("abcd", "abcd"), 0)
def test_mutate(self):
"""mutate with no mutation rate returns the parent."""
alphabet = "abcdefgh"
self.assertEqual(mutate("abcd", 0, alphabet), ['a', 'b', 'c', 'd'])
"""mutate returns only characters from the alphabet."""
self.assertTrue(all([l in alphabet for l in mutate("abcd", 0.25, "abcdefgh")]))
def test_mate(self):
self.assertEqual(mate("abcd", "efgh"), ('abcd', 'efgh', 'abgh', 'efcd'))
def test_evolve(self):
"""evolve raises error if target is not composed of the alphabet."""
with self.assertRaises(AssertionError):
evolve("abcd", "abcx", "abcd")
"""evolve raises error if seed and target are different lengths."""
with self.assertRaises(AssertionError):
evolve("abc", "abcd", "abcd")
print("Evolve some strings.")
"""
Classic example. From random seed to english sentence.
"""
seed = "RHBpoxYLCGjNpUgLYnMfiKskRHmk"
target = "METHINKS IT IS LIKE A WEASEL"
alphabet = "abcdefghijklmnopqrstuvwxyzABCDEFGHIJKLMNOPQRSTUVWXYZ "
generations = evolve(seed, target, alphabet)
print(f"Success in {generations} generations!")
"""
DNA example.
"""
seed = "CGATGATGTATACTGTACGTATCTACTAC"
dna_target = "AATCCGCTAGGTATCAGACTAGTAGCAGT"
dna_alphabet = "ATCG"
dna_generations = evolve(seed, dna_target, dna_alphabet)
print(f"Success in {dna_generations} generations!")
print("Run tests")
unittest.main()- Code is organized into functions with well-defined domains (concerns), and well-defined interfaces.
- Functions are purely functional unless there’s a good reason. Good reasons include needing to throw exceptions, write to logs, generate random numbers, and limited use of global variables.
- The “module” has a public interface (
evolve). If this were a true module in the Python sense, or a class, all other methods could be private. evolvereturns a useful value to the caller. Functions should, in general, return values instead of modifying variables in the body, printing to the screen, writing to files etc. There’s nothing wrong with these things but they are easier to manage at the top level of a program.- All parameters are exposed to the caller. The module doesn’t need to be modified to use the whole space of possible seeds, targets, and population sizes.
5.10 Statistics and other Modeling
Statistics and other forms of modeling and simulation are the most common form of software produced in the practice of science. It is also an area that affects reproducibility and interpretation of research. In a Nature survey assessing the effect of implementing a publication checklist, respondents strongly associated statistical reporting with research quality: “Of those survey respondents who thought the checklist had improved the quality of research at Nature journals, 83% put this down to better reporting of statistics as a result of the checklist.” Nature 556, 273-274 (2018)
[TBD]
5.11 Guidelines for Writing Code
- Write comments. Cover the intention, and how this code fits in with the rest of the codebase, and any meta-information such as deprecation.
- Lint. Use a linter before committing. Git hooks can be set up to run a linter every time you commit. This helps avoid committing a large number of linter corrections at once.
- Write unit tests. Unit tests written early will give you confidence to refactor code, and they help to verify a user is getting the same results.
- Use version control regularly.
- Write code that’s meant to be read and understood.
5.12 Standards
See also Reproducible research
The key qualities that make a codebase reproducible and reusable are:
- Clarity: How simply and understandably the concepts are presented.
- Determinism: How well the starting state of the code, data, and environment are controlled so the user produces the same result as the researcher.
- Interoperability: How well the code interfaces with other tools, and has a standard method for interacting with it.
Document your code, preferably as you go. Even if these are rough notes at first, they are very valuable for reuse.
- Documentation should be tied to a version. The version of the README in a repo should be about the same version of the code.
- Document for superficial users who will only run the runbook and leave, and users who might want to contribute to, or reuse your code.
Do versioned releases. Many times there will only be one release when a paper is published for instance, but it pins your code and documentation to a point in time that is canonical.
For analysis, include a runbook. It should start with data in the same format the user will receive and include any transformations. It should run tests, or give some indication that the run was successful.
Include your data if possible. If your data is not publicly available, direct the users who will have access clearly to the data in the same format and file name they will receive. If the data comes from a database or API, connect to it as simply as possible to avoid confusion.
Include any output data (charts, files) in the repository if possible. This gives the reader a quick reference without running the program, and will alert them if their results are slightly different.
Include dependencies such as libraries, languages, and applications with versions.
Treat the repository as a published artifact. Respect publishing norms such as attribution and long-term access.
5.13 Lab Systems
5.13.1 Version Control
Version control systems such as git are nearly indispensable for maintaining the accuracy and integrity of a codebase. Services like Github also facilitate publishing code in a verifiable, archivable, and contributor-friendly format.
There are eight git commands that are the backbone of git usage. These commands can be memorized in one sitting and provide most of what is needed for the vast majority of routine use.
git clone
# Clone (copy) a repo to the current directory.
git add
# Begin tracking a file or directory. "Add" it to the local repo.
git checkout <branch name>
# Make <branch name> the current branch.
git checkout -b <new branch name>
# Create and check out <new branch name>.
git pull
# Copy the latest versions of the files in the current branch.
git status -s
# Get a list of files that have been modified locally.
git diff
# Get the differences between the local files and the current branch.
git commit -am "<new commit message>"
# Commit (create a set of changed files to be sent to the remote repository).
git push
# Push new commit(s) from the local to the remote repository.
The key to understanding git is 1. Understanding why it is used and 2. A good mental model of repositories, branches, and commits.
Repository: A copy of all the code and history for a particular project. The remote repository is usually on a remote server or service such as github.com.
Branch: A copy of the files in a repository that can be modified independently of other branches. Reconciling the differences between two branches and consolidating them into one branch is merging them.
Commit: A bundle of changes to the files in a repository. The history of a repository consists of a list of commits to various branches, and merges of the branches into another. (Usually there’s one main branch where all the changes eventually end up. The canonical current state of the repo is the state of the main branch, and versioned releases are usually of the main branch.)
5.13.2 Midway
Note: This section was updated in October 2024. At this time the majority of computing takes place on Midway3, but Midway2 is still functional.
Midway2 and Midway3 are the main computing clusters used by the lab, run by the Research Computing Center. Midway3 was launched in March 2021. The official documentation for Midway2 and Midway3 are maintained here by RCC:
Midway2: https://rcc.uchicago.edu/docs/
Midway3: https://mdw3.rcc.uchicago.edu/
This page contains some tasks that are likely to be useful to you.
5.13.2.1 Storage on Midway
There are three places you can store things on Midway: (1) your home directory; (2) your scratch space; and (3) a shared project directory for the lab.
Your home directory is in /home/<CNetID> and has a quota of 30 GB. This is a good place to check out your code, install software you want to use for runs, etc. You should NOT, in general, store simulation output in your home directory.
Your scratch space is /scratch/midway3/<CNet ID>/ and has a quota of 100 GB. This is where you should dump simulation output. I recommend organizing your simulation output using a predictable system: I like to put each experiment in a directory tagged by the date and a name, e.g., 2014-12-08-antigen-vaccine; you may want prefer a hierarchy.
The lab project directory is /project2/cobey for Midway2 (and /project/cobey for Midway3 but as of 8/2021 there is no storage quota) and has a quota of 4.49 TB, with a hard limit of 4.94 TB, for everyone. If you want to share simulation output with other people in the lab, put them here. Keep things organized into separate project directories; I would recommend tagging directories with dates as above, e.g. /project2/cobey/bcellproject-storage/2014-12-08-test. The lab directory has a file quota of 1,885,693 files, with a hard limit of 2,074,262 files.
To check your disk usage, use the quota command. These quotas are current as of August 2021.
5.13.2.2 Connecting to Midway
Details here:
https://rcc.uchicago.edu/docs/connecting/
5.13.2.2.1 SSH (terminal)
To connect via ssh, use your CNetID:
$ ssh <CNetID>@midway2.rcc.uchicago.eduPasswordless login is no longer available, and two factor authentication is required.
5.13.2.2.2 SCP
You can copy individual files or directly back and forth using the scp command, e.g.,
$ scp <local-path> <CNetID>@midway2.rcc.uchicago.edu:<remote-path>
$ scp <CNetID>@midway2.rcc.uchicago.edu:<remote-path> <local-path>There are also graphical SSH/SCP browsers for Mac OS X, Linux, and Windows:
5.13.2.2.3 SAMBA (SMB) (connect as a disk)
You can make Midway to look like a local disk on your computer using SAMBA. This is convenient for things like editing job submission scripts using your favorite editor directly on the server, without having to copy things back and forth.
Midway’s SAMBA (SMB) hostname is midwaysmb.rcc.uchicago.edu.
If you’re off campus, you’ll need to be connected to the U of C VPN to access SMB. It will also be pretty slow, especially if you’re on a crappy cafe connection like the one I’m on now–you may find a GUI SCP client to be a better choice offsite.
On Mac OS X, go to the Finder, choose Go > Connect to Server… (Command-K), and then type in the URL for the directory you want to access. The URLs are currently confusing (you need to specify your CNetID in the scratch URL but not in the home URL):
Home: smb://midwaysmb.rcc.uchicago.edu/homes
Scratch: smb://midwaysmb.rcc.uchicago.edu/midway-scratch/<CNetID>
Project: smb://midwaysmb.rcc.uchicago.edu/project/cobey5.13.2.2.4 ssh -X (terminal with graphical forwarding)
If you connect to Midway via ssh -X, graphical windows will get forwarded to your local machine. On Linux, this should work out of the box; on Mac OS X you’ll need to install XQuartz first. (There’s probably a way to make this work on Windows too; if you figure this out, please add instructions here.)
It’s then as simple as doing this when logging in:
$ ssh -X <CNetID>@midway2.rcc.uchicago.eduThis is especially useful for the sview command (see below); it also will forward the graphics of a job running on a cluster node if you use ssh -X and thensinteractive`.
5.13.2.2.5 VNC (graphical interface via ThinLinc)
You can also get a full Linux desktop GUI on a Midway connection using a program called ThinLinc:
https://rcc.uchicago.edu/docs/connecting/#connecting-with-thinlinc
WARNING: the first time you use ThinLinc, before you click Connect go to Options… > Screen and turn off full-screen mode. Otherwise ThinLinc will take over all your screens, making it rather hard to use your computer.
5.13.2.2.6 Connecting to Eduroam internationally
See UChicago ServiceNow.
5.13.2.3 Configuring Software on Midway
Before you run anything on Midway, you’ll need to load the necessary software modules using the module command:
https://rcc.uchicago.edu/docs/tutorials/intro-to-software-modules/
If you’re not sure what the name of your module is, use, e.g.,
$ module avail inteland you’ll be presented with a list of options. You can then load/unload modules using, e.g.,
$ module load intel/15.0
$ module unload intel/15.0There are multiple versions of many modules, so you’ll generally want to check module avail before trying module load.
If you want to automatically load modules every time you log in, you can add module load commands to the end of your ~/.bash_profile file (before # User specific environment and startup programs).
5.13.2.5 Java
$ module avail javaAs of writing, Midway only has one version of Java (1.7), so be sure not to use JDK 1.8 features in your Java code.
5.13.2.6 C/C++
$ module avail intel
$ module avail gccThere are three C/C++ compiler modules available on Midway: gcc, intel, and pgi. The Intel and PGI compilers are high-performance compilers that should produce faster machine code than GCC, but only intel seems to be kept up to date on Midway, so I’d recommend using that one.
The Intel compilers should work essentially the same as GCC, except due to ambiguities in the C++ language specification you may sometimes find that code that worked on GCC needs adjustment for Intel. To use the Intel compiler, just load the module and compile C code using icc and C++ code using icpc.
Unless you have a good reason not to, you should use module avail to make sure you’re using the latest GCC and Intel compilers, especially since adoption of the C++11 language standard by compilers has been relatively recent at the time of writing.
Also, it’s worth knowing that Mac OS X, by default, uses a different compiler entirely: LLVM/Clang. (Currently, only an old version of LLVM/Clang is available on Midway). So you might find yourself making sure your code can compile using three different implementations of C++, each with their own quirks.
Here’s how you compile C++11 code using the three compilers:
$ g++ -std=c++11 -O3 my_program.cpp -o my_program
$ icpc -std=c++11 -O3 my_program.cpp -o my_program
$ clang++ -std=c++11 -stdlib=libc++ -O3 my_program.cpp -o my_programNOTE: the -O3 flag means “optimization level 3”, which means, “make really fast code.” If you’re using a debugger, you’ll want to leave this flag off so the debugger can figure out where it is in your code. If you’re trying to generate results quickly, you’ll want to include this flag. (If you’re using Apple Xcode, having the flag off and on roughly correspond to “Debug” and “Release” configurations that you can choose in “Edit Scheme”.)
If you want to make it easy to compile your code with different compilers on different systems, you can use the make.py script in the bcellmodel project as a starting point. It tries Intel, then Clang, then GCC until one is available. (This kind of thing is possible, but a bit annoying to get working, using traditional Makefiles, so I’ve switched to using simple Python build scripts for simple code.)
5.13.2.7 R
$ module avail Rshows several versions of R. The default version (as of August 2021) is R/3.6.1. The best version will probably be the latest one alongside the Intel compiler, e.g., R/3.3+intel-16.0 at the time of writing. You can choose which version of R to use:
$ module load R/4.0.1If you have already loaded the default version of R ($ module load R), you will need to first unload R ($ module unload R) before you can specify a version.
5.13.2.8 Python
In order to keep a consistent Python environment between your personal machine and Midway, we are maintaining our own Python installations in /project/cobey. Skip the Midway Python modules entirely, and instead include this in your ~/.bash_profile:
export PATH=/project/cobey/anaconda/bin:/project/cobey/pypy/bin:$PATHTo make sure things are set up properly make sure that which python finds /project/cobey/anaconda/bin/python and which pypy finds /project/cobey/pypy/bin/pypy. See the Python page on this wiki for more information.
5.13.2.9 Matlab and Mathematica
You shouldn’t use Matlab or Mathematica if possible, because if you publish your code your results will only be reproducible to people that want to pay for Matlab or Mathematica.
But if you must…
module avail matlab
module avail mathematicashows that they are available on Midway2. (Getting a graphical Mathematica notebook to run on the cluster is a pain in the ass, though, so you’re probably better off just running it locally if you can get away with it.)
5.13.2.10 Installing R and Python libraries
5.13.2.10.1 R
We usually need specific libraries to run R scripts. It is important to check what version of R are necessaryfor libraries to be installed and adjust accordingly. I find it helpful to create an installation.R script to install libraries into the correct directory, for example:
#!/usr/bin/Rscript
## Create the personal library if it doesn't exist. Ignore a warning if the directory already exists.
dir.create(Sys.getenv("R_LIBS_USER"), showWarnings = FALSE, recursive = TRUE)
## Install multiple packages
install.packages(c('dplyr',
'optparse',
'doParallel'),
dependencies = T,
Sys.getenv("R_LIBS_USER"),
repos = 'http://cran.us.r-project.org'
)Other libraries (such as panelPomp), are not yet in the CRAN respository. These libraries often need to be installed from GitHub, which is possible using devtools.
## panelPomp from GitHub
install.packages(c("devtools"),
dependencies = T,
Sys.getenv("R_LIBS_USER"),
repos = 'http://cran.us.r-project.org'
)
devtools::install_github('cbreto/panelPomp')5.13.3 Running Jobs on Midway
I’ll leave most of the details to the official documentation:
https://rcc.uchicago.edu/docs/using-midway/#using-midway
but a summary of important stuff follows.
5.13.3.1 Overview
Midway consists of a large number of multi-core nodes, and uses a system called SLURM to allocate jobs to cores on nodes.
Some details on the terminology: a “node” is what you normally think of as an individual computer: a box with a motherboard, a hard drive, etc., running a copy of Linux. Each node’s motherboard contains several processors (a physical chip that plugs into the motherboard), each of which may contain several cores. A “core” is a collection of circuits inside the processor that can, conceptually speaking, perform one series of instructions at a time. (Until a few years ago, processors only consisted of one core and people talked about “processors” the way they now talk about “cores,” so you might hear people confusing these from time to time.)
If you are writing code that does only one thing at a time (serial code), then all you really need to know is that a single run of your code requires a single core.
5.13.3.2 Job structure
Note that Midway counts service units, or core-hours in increments of 0.01. To minimize waste, we’re best off designing jobs to be at least several minutes each. (It makes sense that we wouldn’t want to bog down the scheduler anyway.)
5.13.3.3 Priority
As described below, if your jobs aren’t running right away, you can use
$ squeue -u <CNetID>to see what’s going on.
If your jobs are queued with (Priority) status, it means other jobs are taking priority. Job priorities are determined by the size of the job, its time in the queue, the requested wall time (so it pays to be precise and know your jobs well), and group-level prior usage. Groups that have consumed fewer resources get higher priority than those using more. This usage has a half-life of approximately 14 days, which means it’s less awkward to spread jobs out over time. You can view the priority of queued jobs using
$ sprio5.13.3.4 Useful commands
5.13.3.4.1 sinteractive
To get a dedicated job that you can interact with just like any login session–e.g., if you want to manually type commands at the command line to try some code out, make changes, do some analysis, etc.–you can use the sinteractive command:
https://rcc.uchicago.edu/docs/using-midway/#sinteractive
If you connected via ssh -X, then graphical windows will also get forwarded from the cluster to your local machine.
5.13.3.4.2 sbatch
To submit a single non-interactive job to the cluster, use the sbatch command:
https://rcc.uchicago.edu/docs/using-midway/#sbatch
This involves preparing a script with special indications to SLURM regarding how much memory you need, how many cores, how long you want the job to run, etc.
5.13.3.5 scontrol update
Useful for changing the resources requested for PENDING jobs.
Move a PENDING job to another partition:
$ scontrol update partition=<partition_name> qos=<partition_name> jobid=<jobid>Change the time limit for a PENDING job:
$ scontrol update TimeLimit=<HH:MM:SS> jobid=<jobid>5.13.3.5.1 accounts
Display number of used/available CPU-hours for the lab:
$ accounts balanceDisplay number of CPU hours used by you:
$ accounts usageDisplace account usage (SUs) for a previous period (update –cycle to indicate the academic cycle of interest):
$ /software/systool/bin/accounts balance --account=pi-cobey --cycle=2019-20205.13.3.5.2 sview
If you’re connected graphically to Midway (either via ssh -X or using ThinLinc), you can get a graphical view of the SLURM cluster, which makes it easy to do things like selectively cancel a bunch of jobs at once:
$ sviewThe most useful command: Actions > Search > Specific User’s Job(s)
5.13.3.5.3 quota
Display storage usage:
$ quotaThe standard quota on an individual account is 30GB. When you exceed this, Midway will notify you the next time you log in. There is a grace period of 7 days to adjust your usage. There is also a hard limit of 35GB.
The check the file and storage breakdown on \project2\cobey (or \project\cobey), add the -F flag:
$ quota -F /project2/cobey5.13.4 SLURM tricks
squeue -o lets you specify additional information for squeue using a format string. These are annoying to type every time you want to query things. You can create an alias in your .bash_profile script:
which includes standard info plus some extra stuff (including time limit, # nodes, # CPUs, and memory). Then you can just type sq, sq -p cobey, sq -u edbaskerville, etc. to perform queries with your customized format string.
See man squeue for a list of format string options.
sinfo -o has similar options, including the ability to see how many processors are available/in use:
A full list of aliases for user edbaskerville might look like:
5.13.5 Other Midway items to keep track of
5.13.5.1 Protected health information (PHI) and MidwayR
There are instances where we work with data that qualifies as PHI. Most often, this is individual-level data that includes a specific date related to an individual (e.g., birth date, admission date, vaccination date). These types of data cannot be stored or used in jobs running on Midway2/Midway3. For PHI and other protected data types, MidwayR may be useful. MidwayR is similar to the Midway computing environment but is also equipped with tools and software needed to meet the highest levels of secure data protection. Even with using MidwayR, it may be necessary to obscure dates that qualify as PHI.
5.13.5.2 Allocation requests
Each September we have to submit an allocation request to the RCC to request Service Units (SUs). SUs are the basic unit of computational resources and they represents the use of one core for one hour on the Midway Cluster. Allocation requests from 2019-2020 and 2020-2021 are housed on /project2/cobey/allocation_requests_midway_resources.
5.13.5.3 Node life
For each node purchased by the lab, the warranty is 5 years. Once the warranty runs out, the nodes can still be active on Midway2, but if they break or go down, the RCC will not fix them. For any long-duration jobs, please ensure you are using a node that is under warranty to reduce the likelihood of data loss. A spreadsheet with nodes, purchase date, warranty duration, and node label is housed in /project2/cobey/allocation_requests_midway_resources.
For the current status of private nodes, use:
sinfo -p cobey
Or for nodes with “cobey” in the name:
sinfo | grep cobey
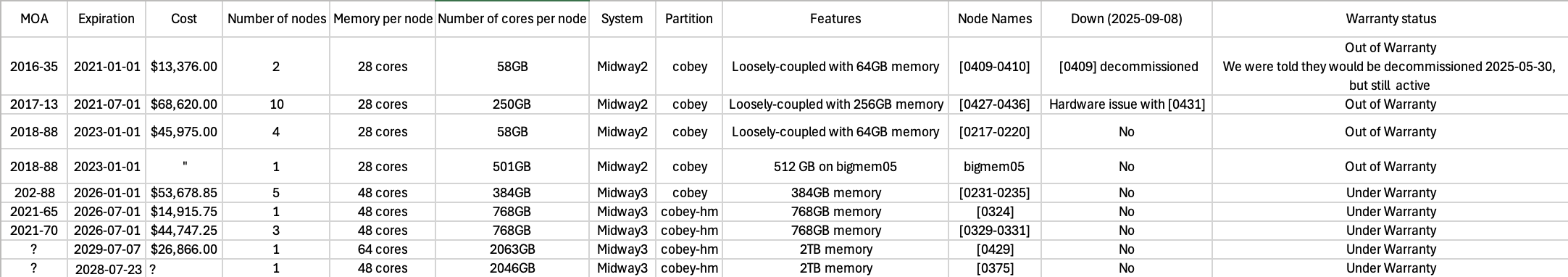
Figure 5.1: Node list with status September 2025
5.13.5.4 Transitioning from Midway2 to Midway3
Any nodes purchased after March 2021 are housed on Midway3. The RCC will mount storage from Midway2 onto Midway3, meaning it will be accessible when running jobs from Midway3. As of August 2021, this has not yet occurred (due to delays related to the pandemic), but they expect this will occurr by October 2021. After this point, it will make sense for all new projects to be housed on Midway3 instead of Midway2.
5.14 PHI and PII
Protected Health Information (PHI) and (PII) must be handled carefully according to the IRB protocol of the project, legal restrictions, and UChicago policy. Policies are covered in the Human Subjects, and HIPAA trainings.
5.14.1 Links to Policies
(Available only within UCM network or VPN)
Responsibilities and Oversight Policy
Personal Computing Device Policy
Data Classification Policy and Handling Procedures
5.14.2 Handling PHI
Data is vulnerable “at rest” (on a local machine, or cloud storage) and “in transit” between machines over the internet or other network. It can also be recovered after it has been deleted from certain types of drive, and copies are often retained on systems where data was stored. Online services (Slack, email systems, Github, messaging apps) – even when attachments appear to be limited to two parties – also expose data to the company that owns the service, and their internal logs, backups, third-party services et cetera.
It is important, therefore, to use secure systems even for temporary storage, or when transferring files to MidwayR. PHI generally needs to be deleted after the conclusion of a study according to a Data Use Agreement, or IRB protocol.
CrashPlan Pro should be set up to exclude any PHI data directories, and backups should be included when deleting data. Copies can also be in recycle bins, logs, and swap files.
There’s no official secure deletion application for individual files. However, CCleaner and similar programs can help with the process on local machines. Whole-disk deletion should be coordinated with the BSD Information Security Office.
If data is exposed, lost or stolen, it should be reported according to the corresponding protocol or agreement.
Methods for transferring files are available in the MidwayR User Guide.